ABOUT
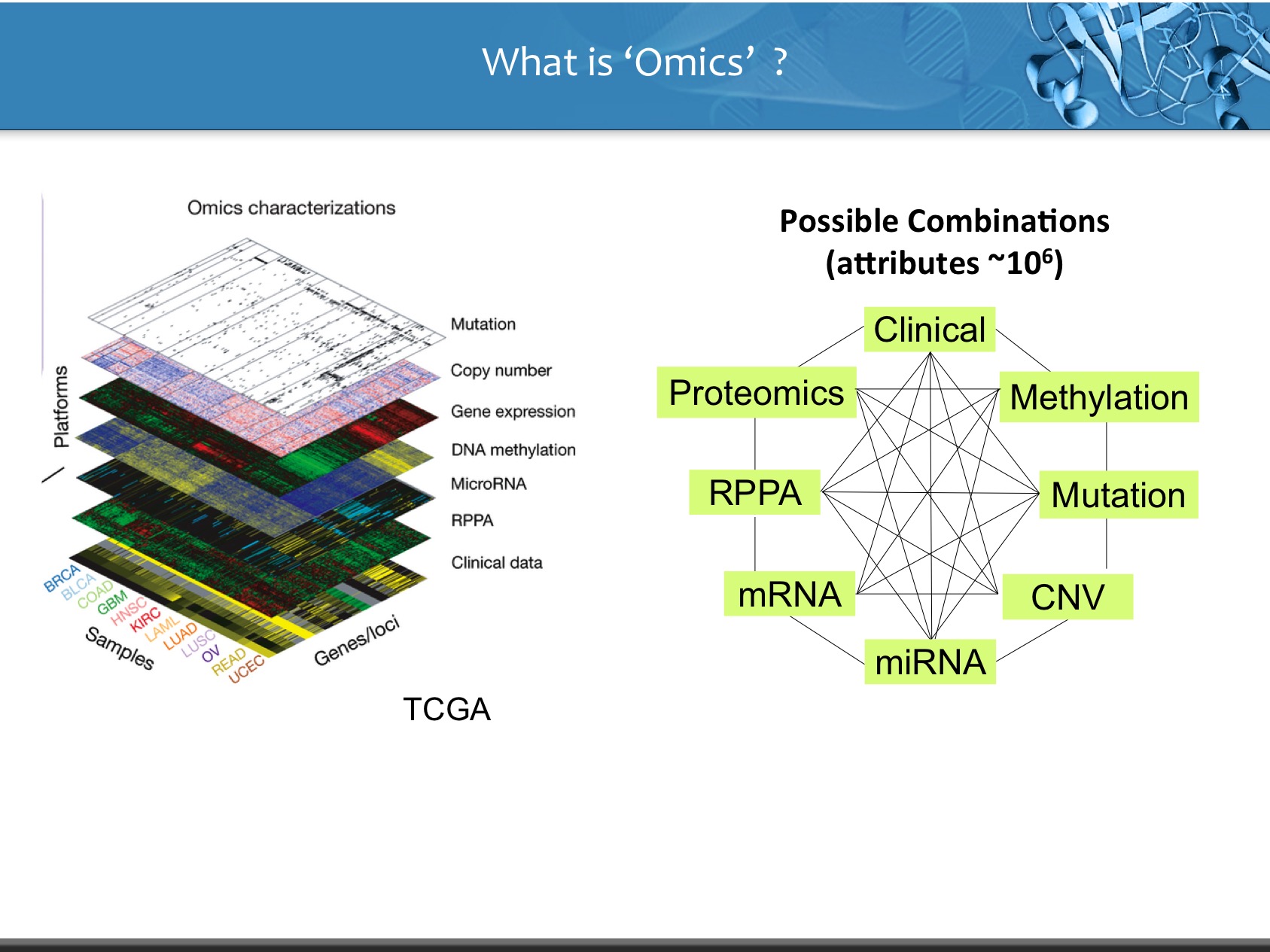
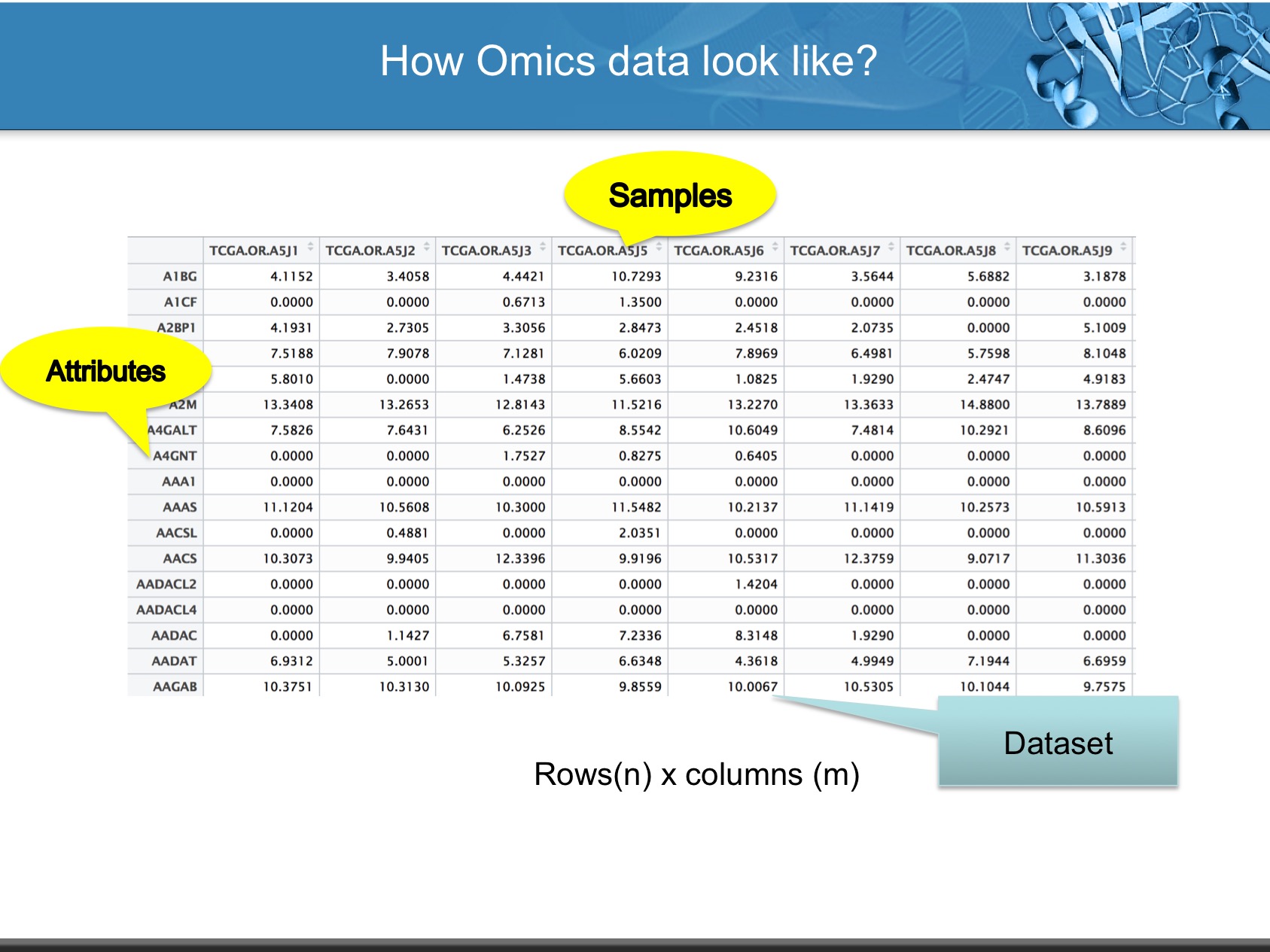
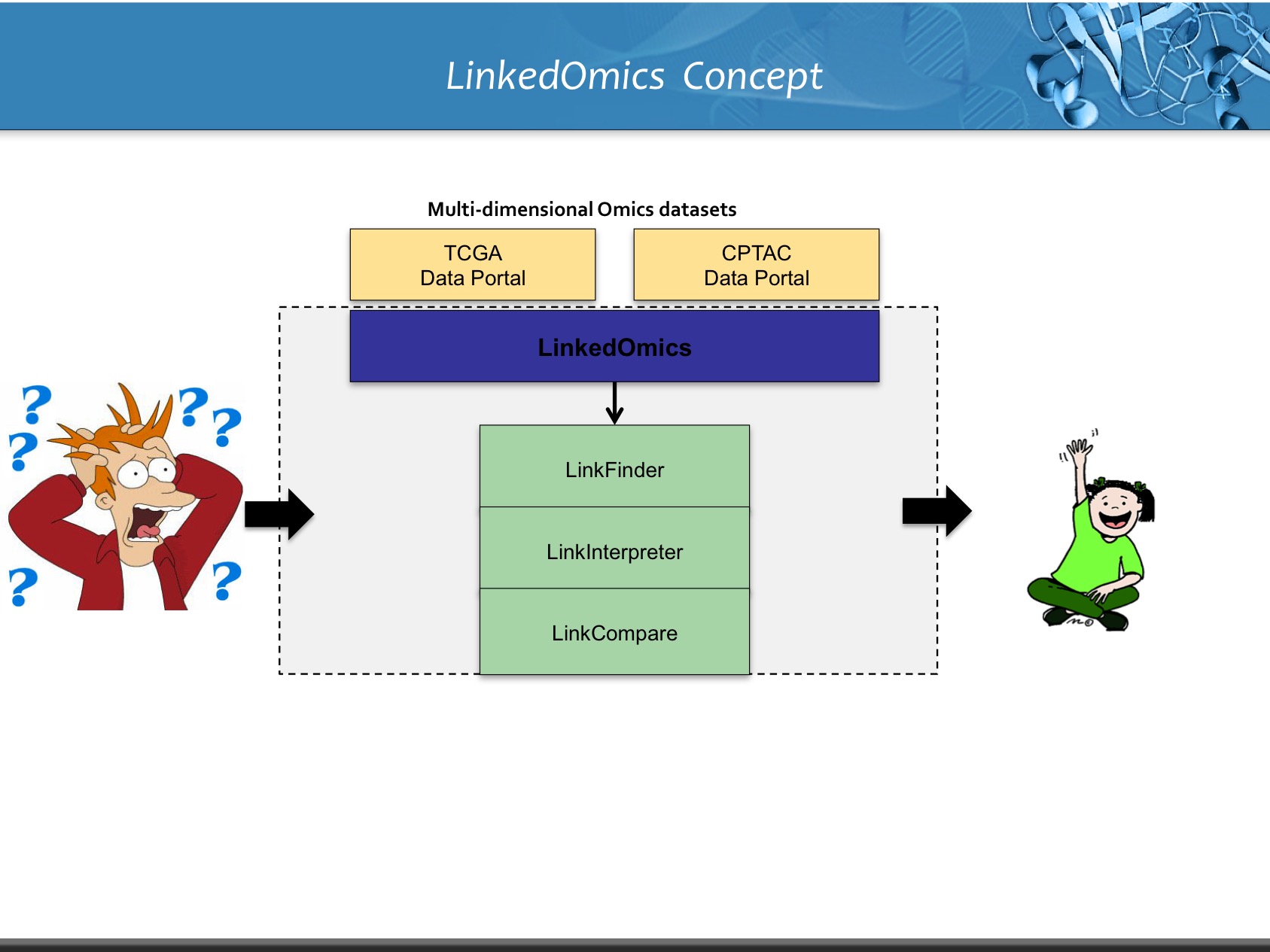
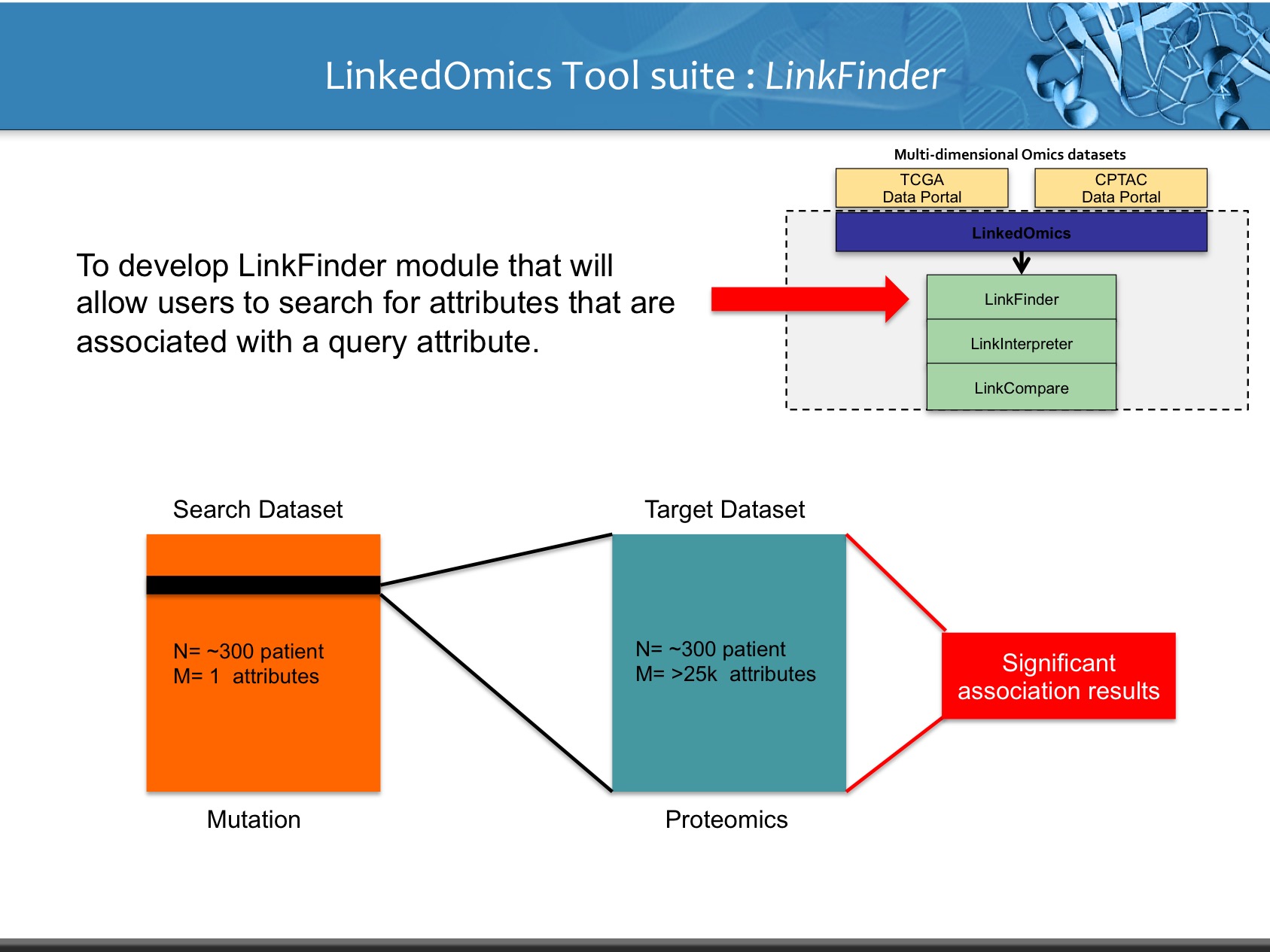
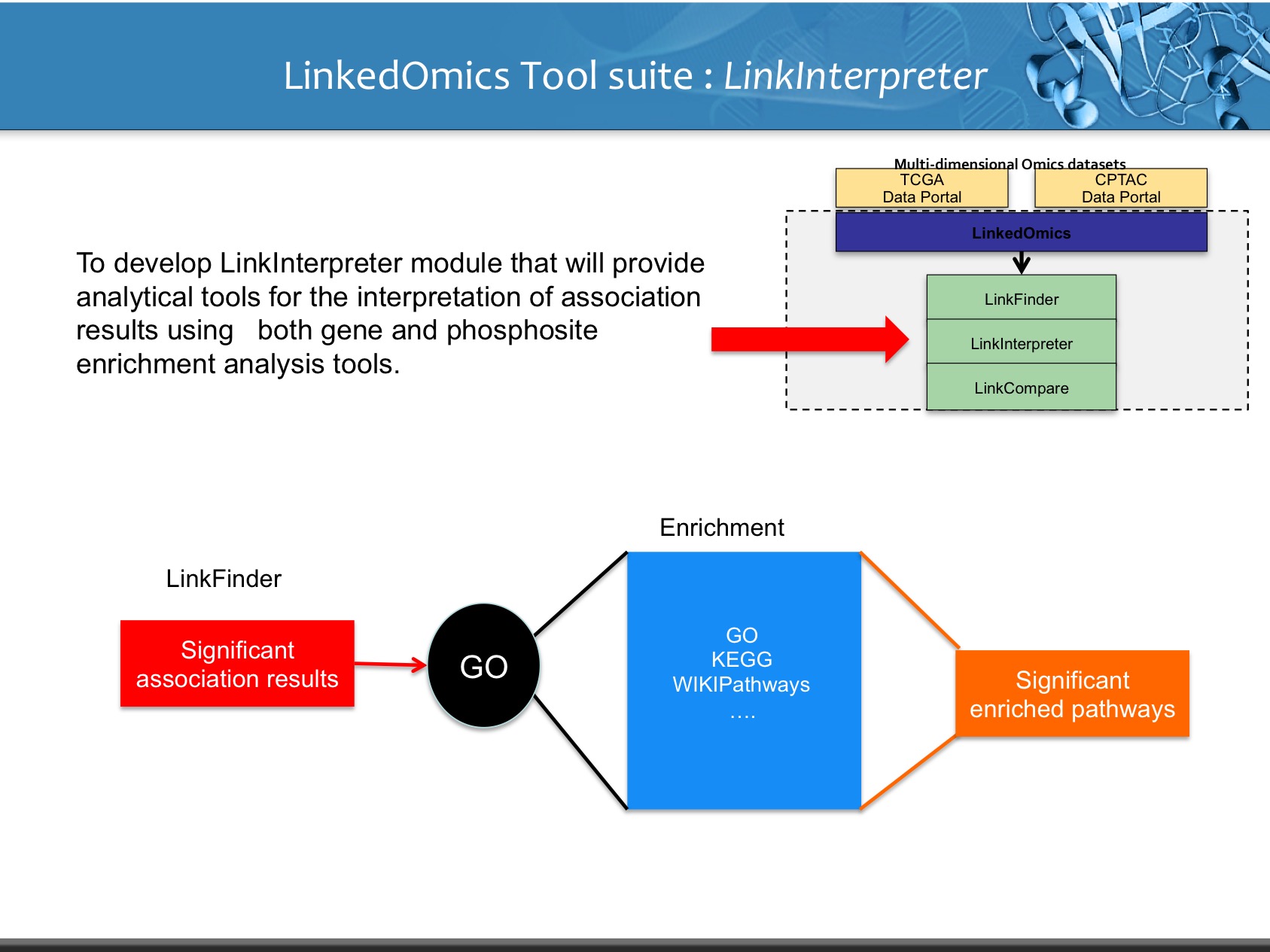
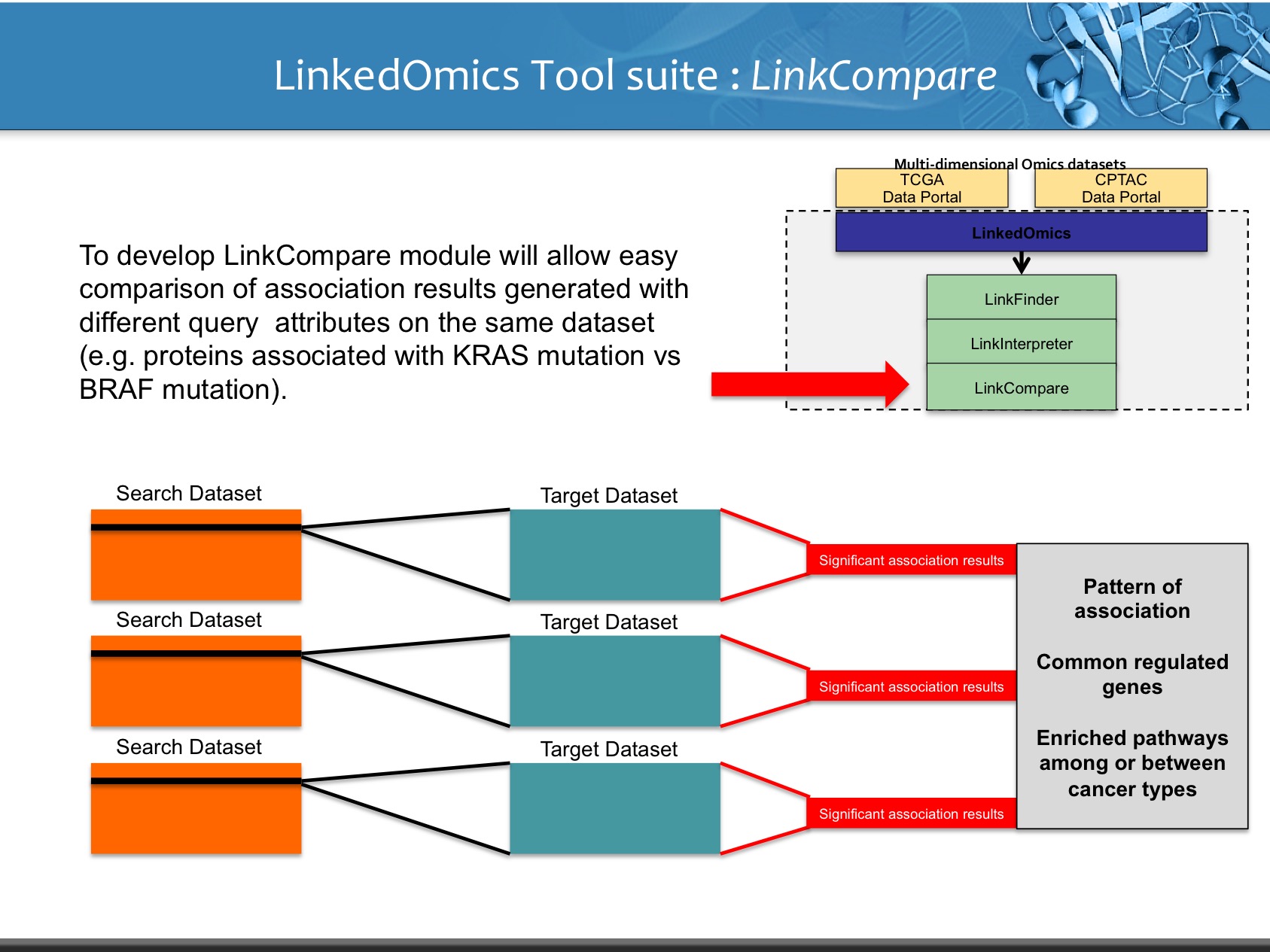
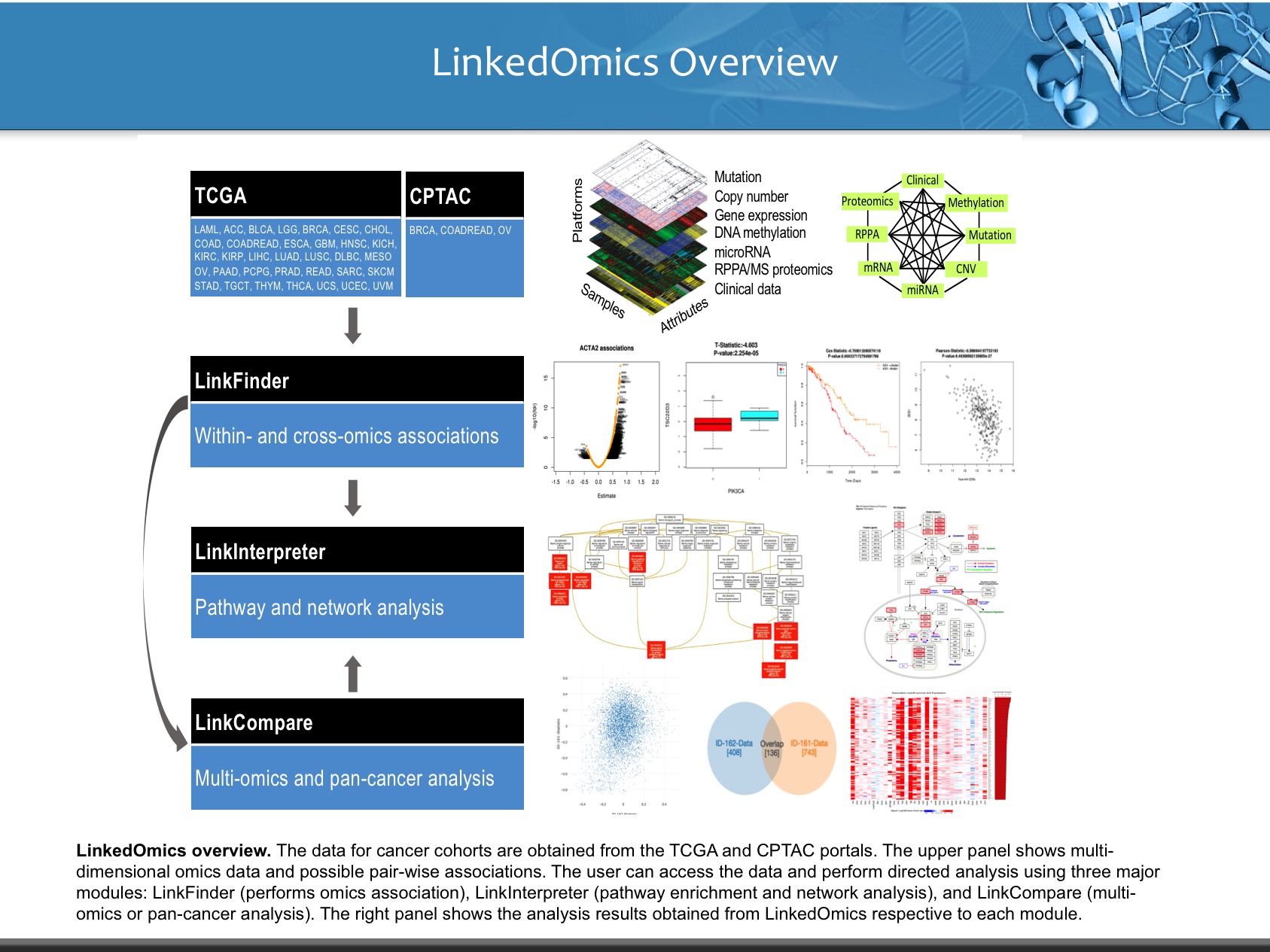
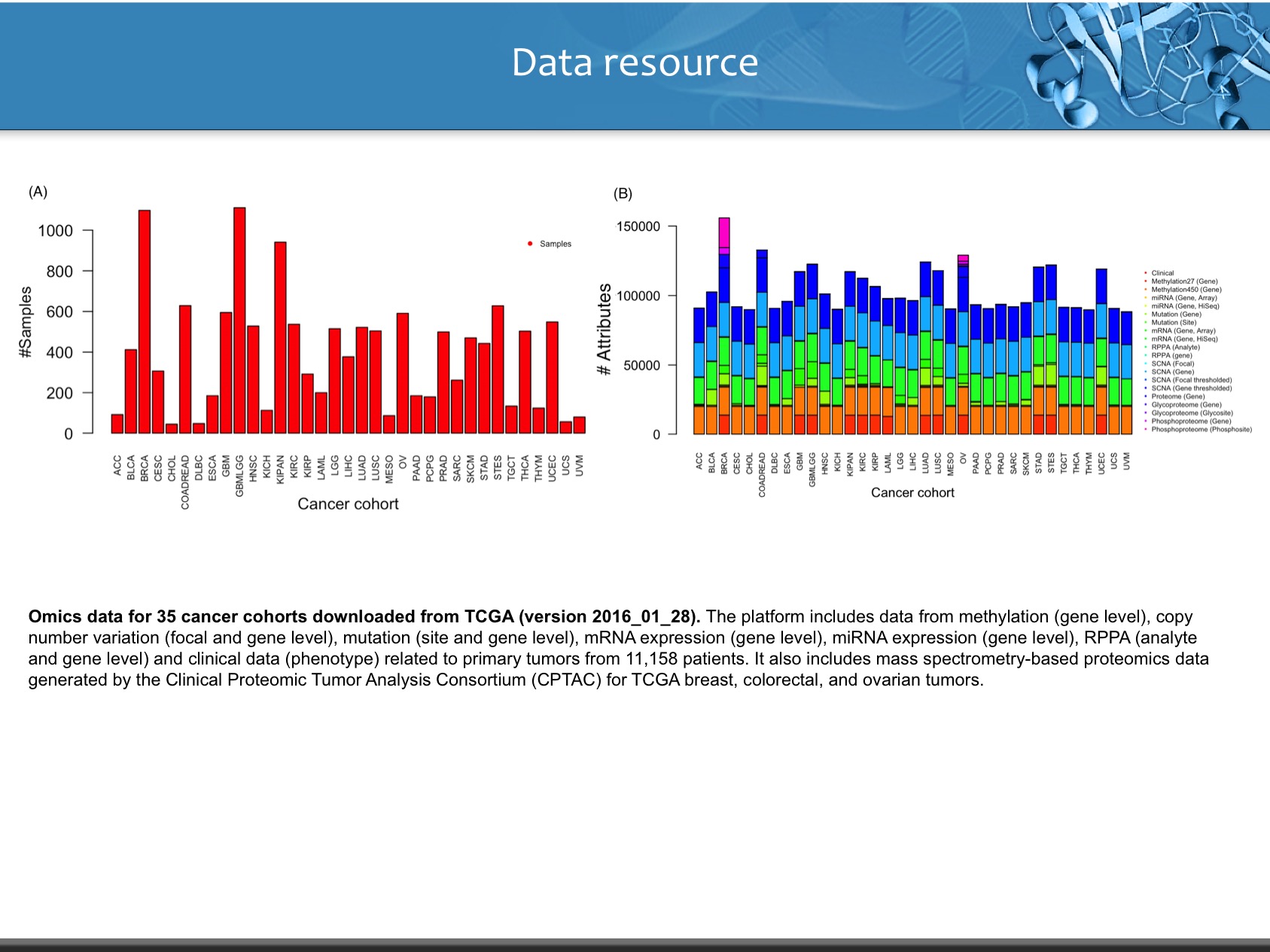
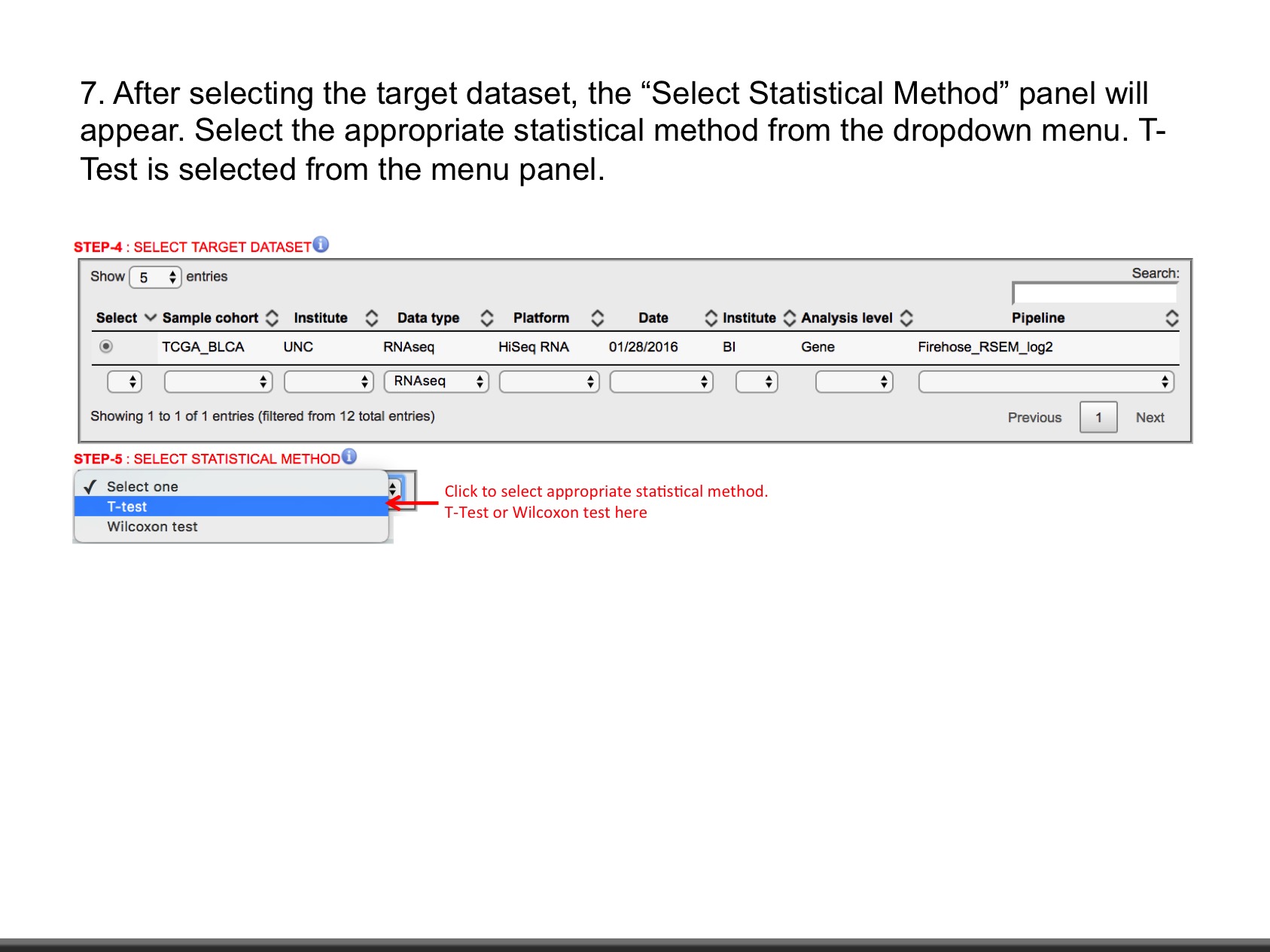
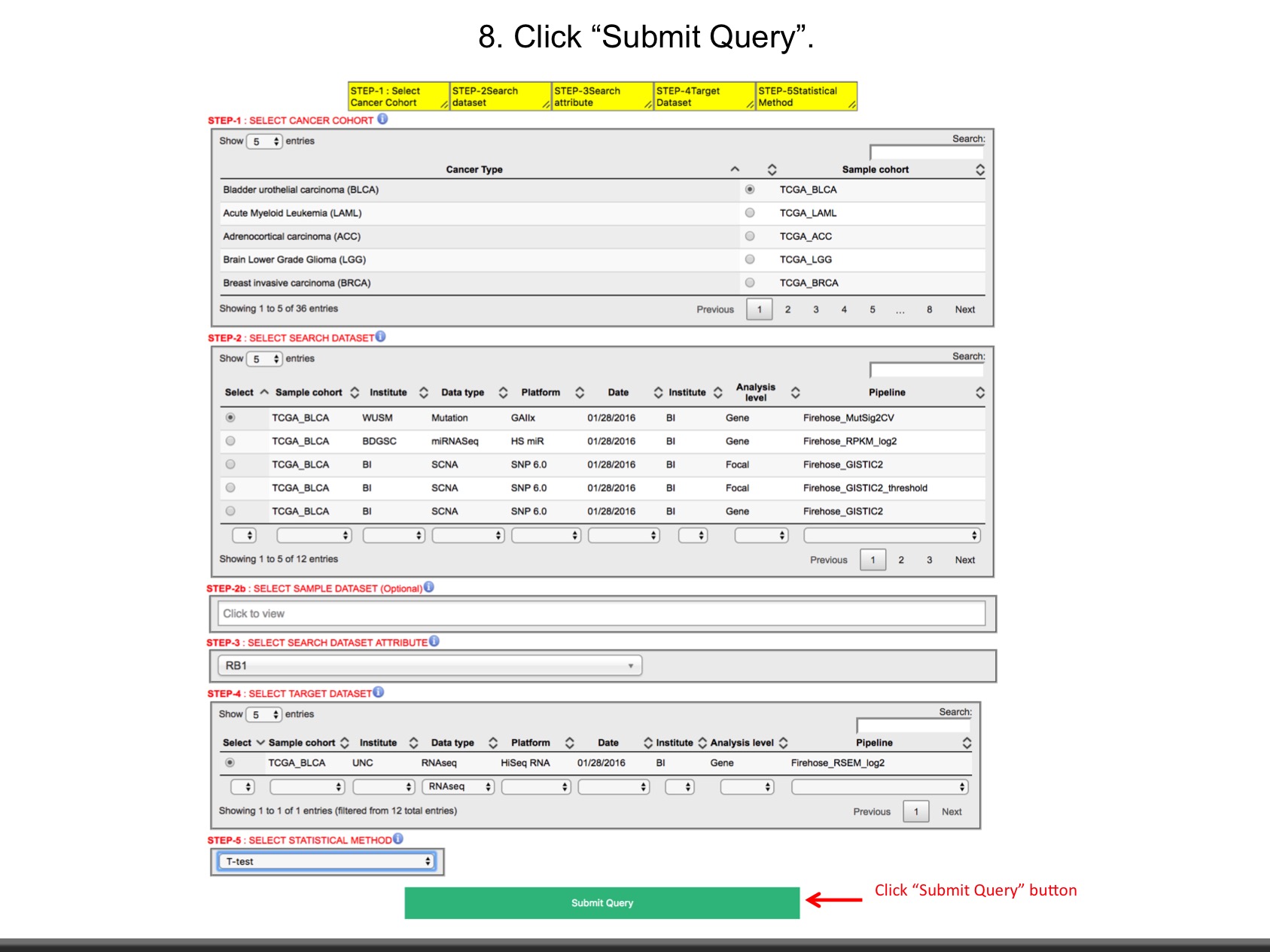
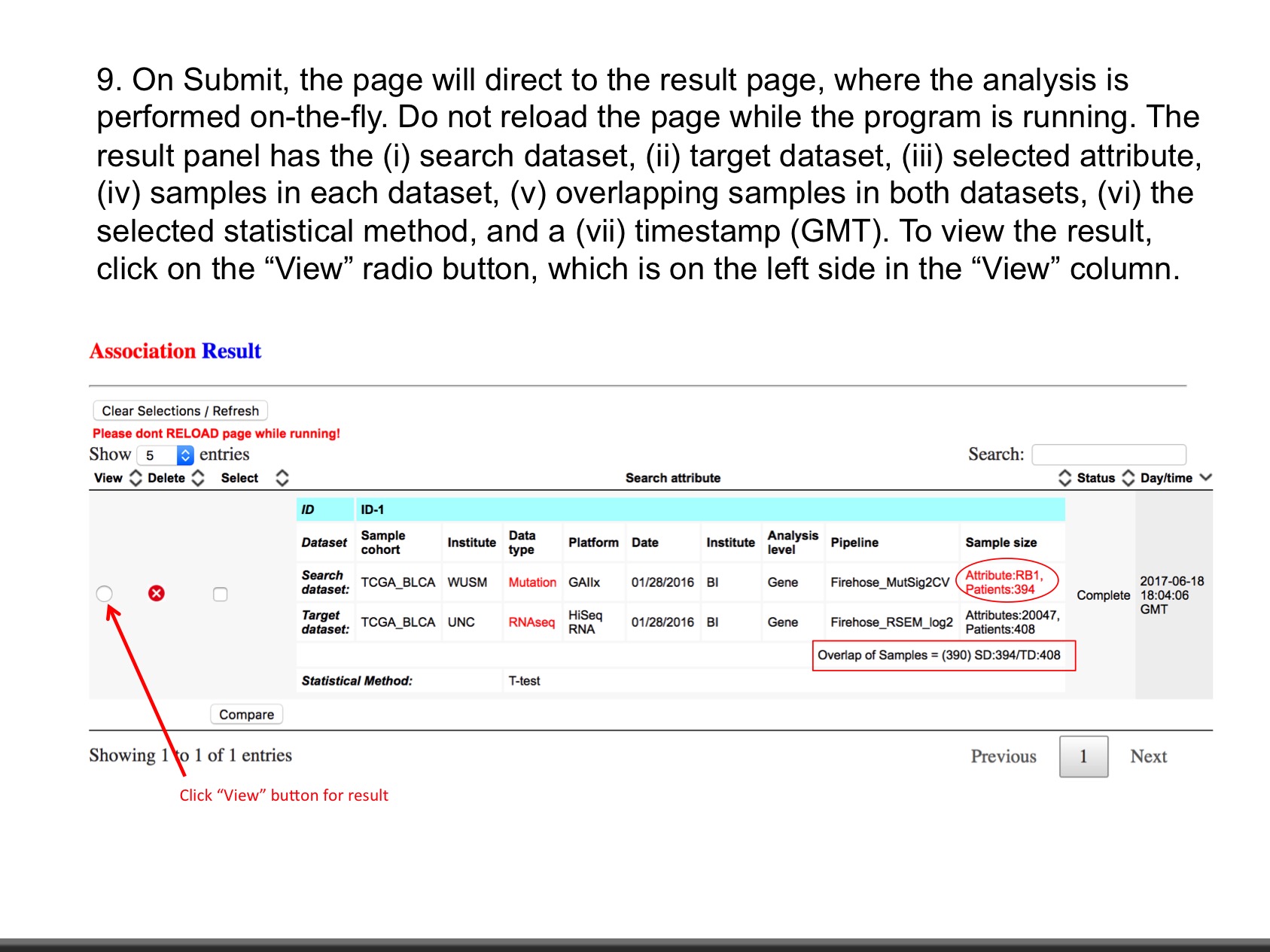
LinkedOmics is publicly available portal that includes multi-omics data from all 32 TCGA tumor types. It also includes mass spectrometry-based proteomics data generated by the Clinical Proteomics Tumor Analysis Consortium (CPTAC) for TCGA breast, colorectal and ovarian tumors. The web application has three analytical modules: LinkFinder, LinkInterpreter and LinkCompare. LinkFinder allows users to search for attributes that are associated with a query attribute, such as mRNA or protein expression signatures of genomic alterations, candidate biomarkers of clinical attributes, and candidate target genes of transcriptional factors, microRNAs, or protein kinases. Analysis results can be visualized by scatter plots, box plots, or Kaplan-Meier plots. To derive biological insights from the association results, the LinkInterpreter module performs enrichment analysis based on Gene Ontology, biological pathways, network modules, among other functional categories. The LinkCompare module uses visualization functions (interactive venn diagram, scatter plot, and sortable heat map) and meta-analysis to compare and integrate association results generated by the LinkFinder module, which supports multi-omics analysis in one cancer type or pan-cancer analysis.
LinkedOmics provides a unique platform for biologists and clinicians to access, analyze and compare cancer multi-omics data within and across tumor types.
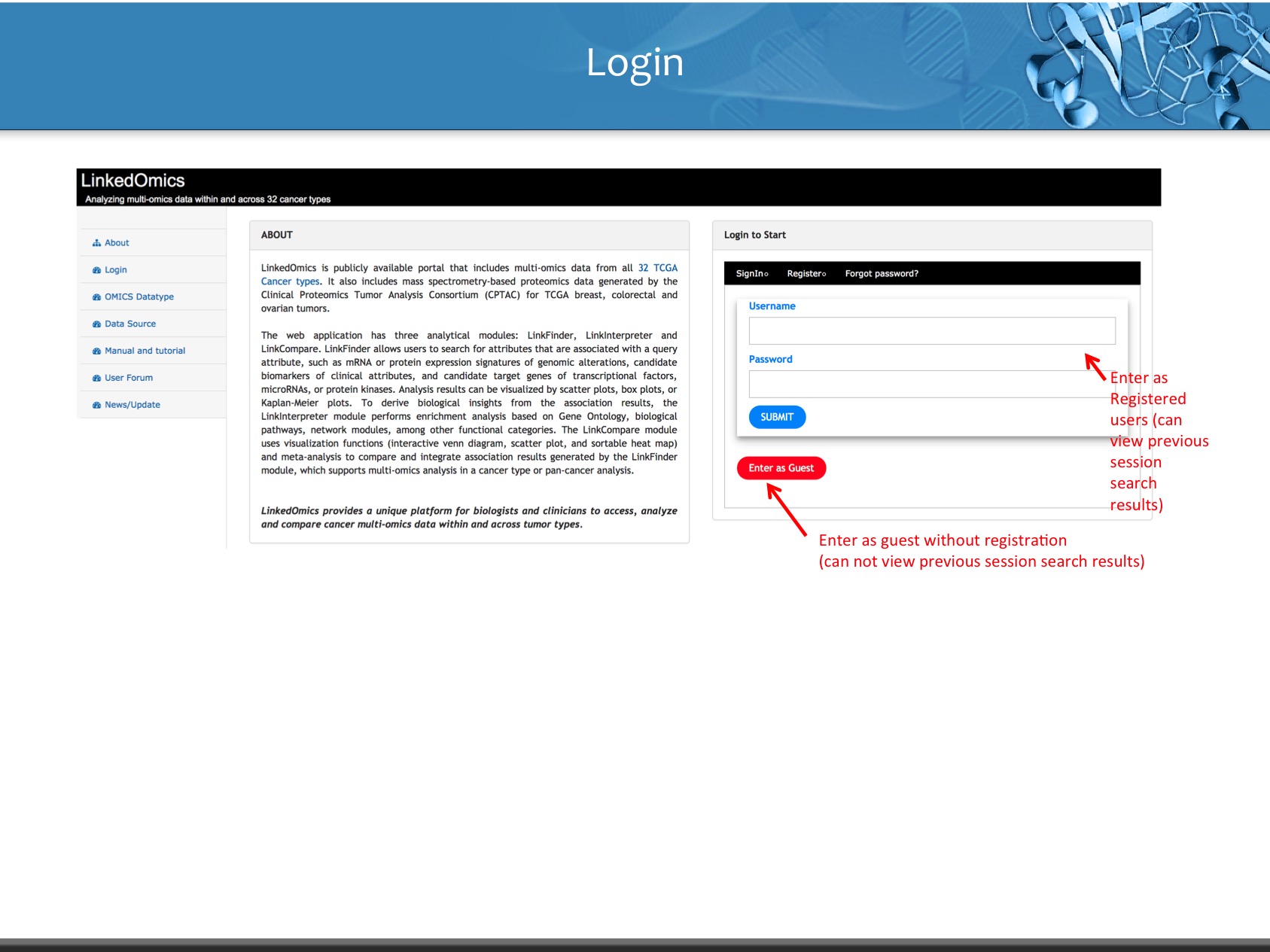
Login to Start
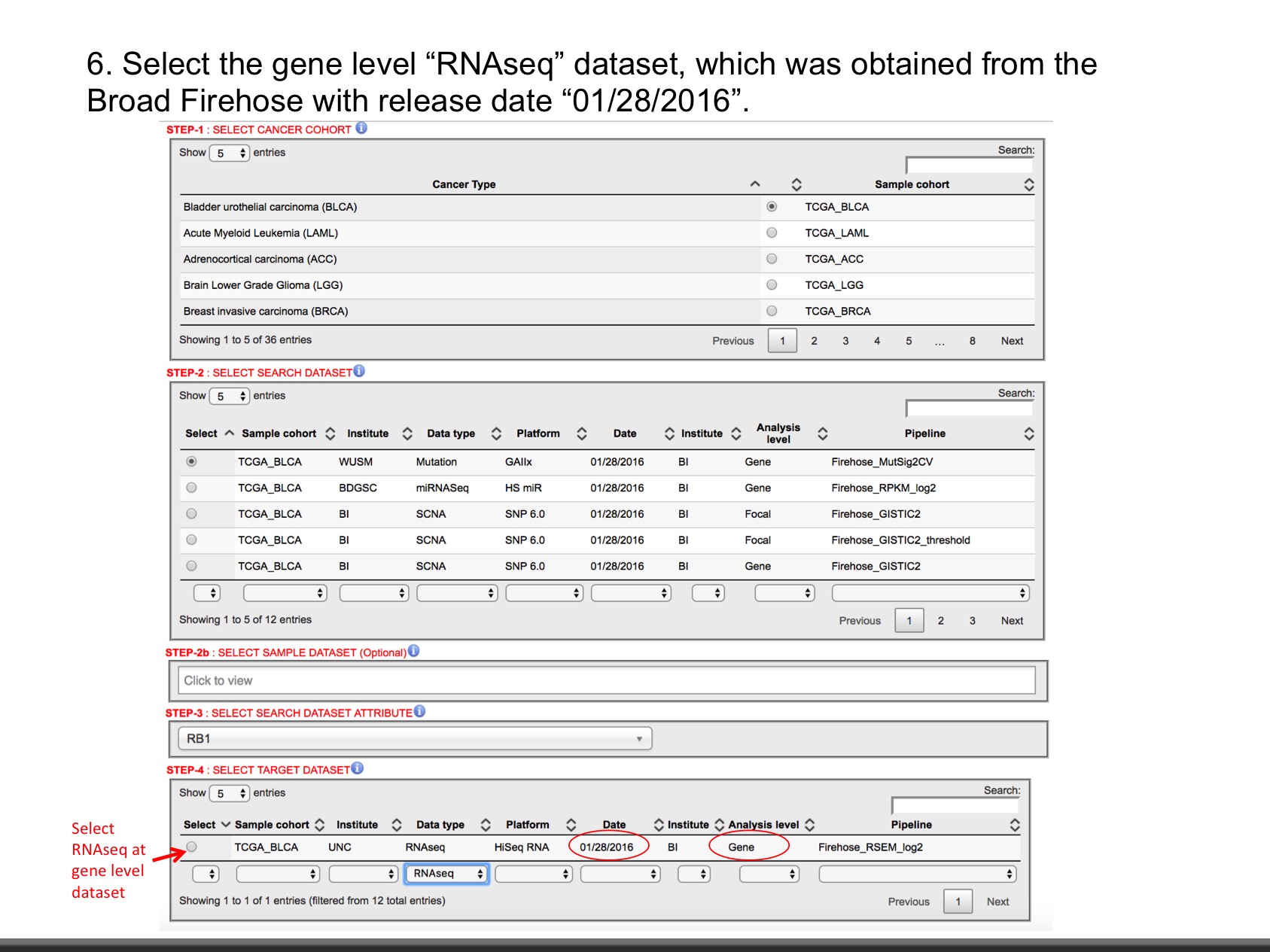
- Clinical Data : It inculdes attributes like age, overall survival, phological stage (I, II, III, IV), TNM staging, Clinical subtype, Molecular Subtype, number of lymph nodes, radiation therapy.
- Copy Number (Level: Focal, Gene) : Normalized copy number (SNPs) ans Copy number alterations for aggregated/segmented regions, per sample
- miRNA (Leve: Gene, isoform) : Normalized signals per probe or probe set for each participant's tumor sample
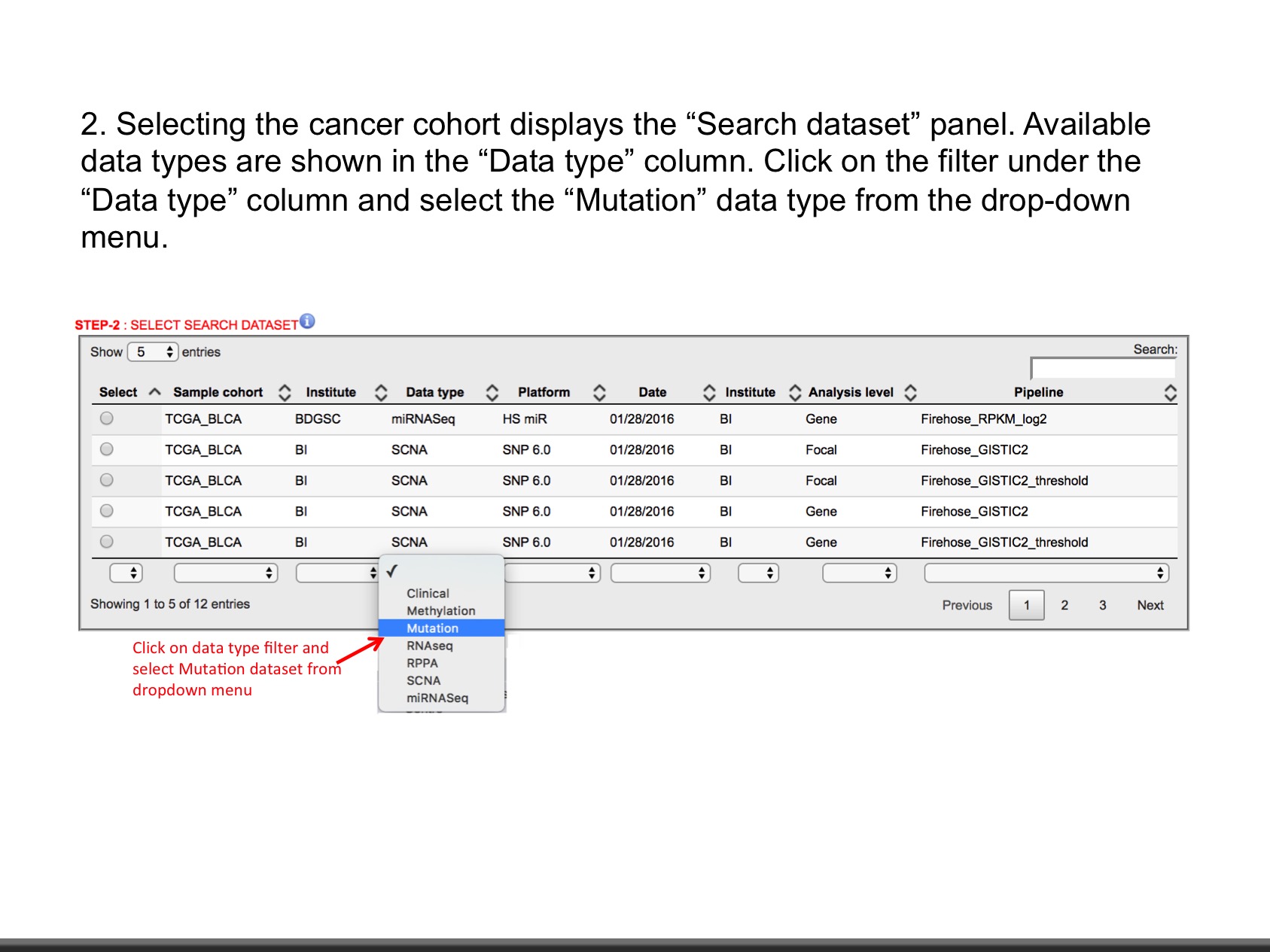
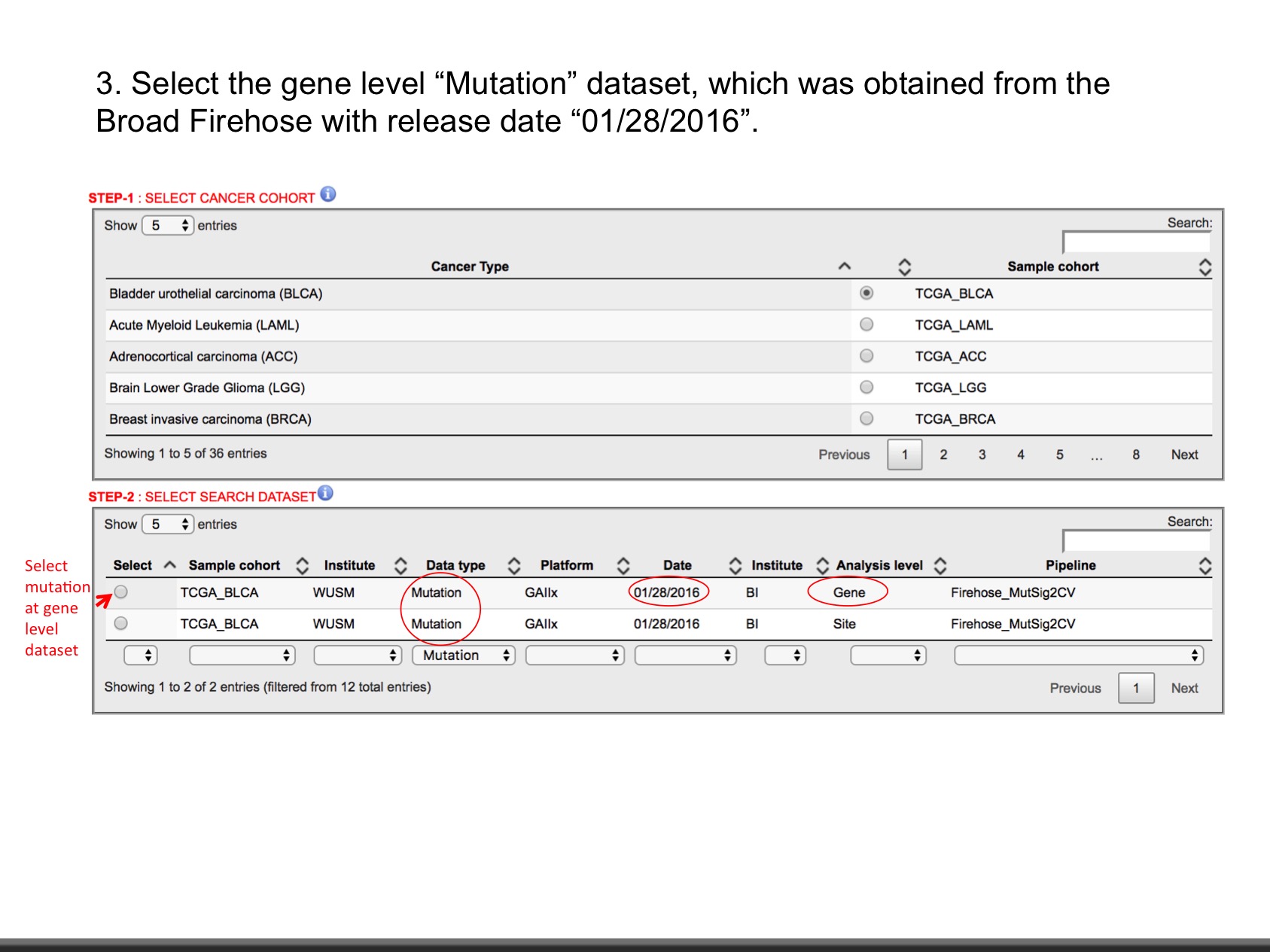
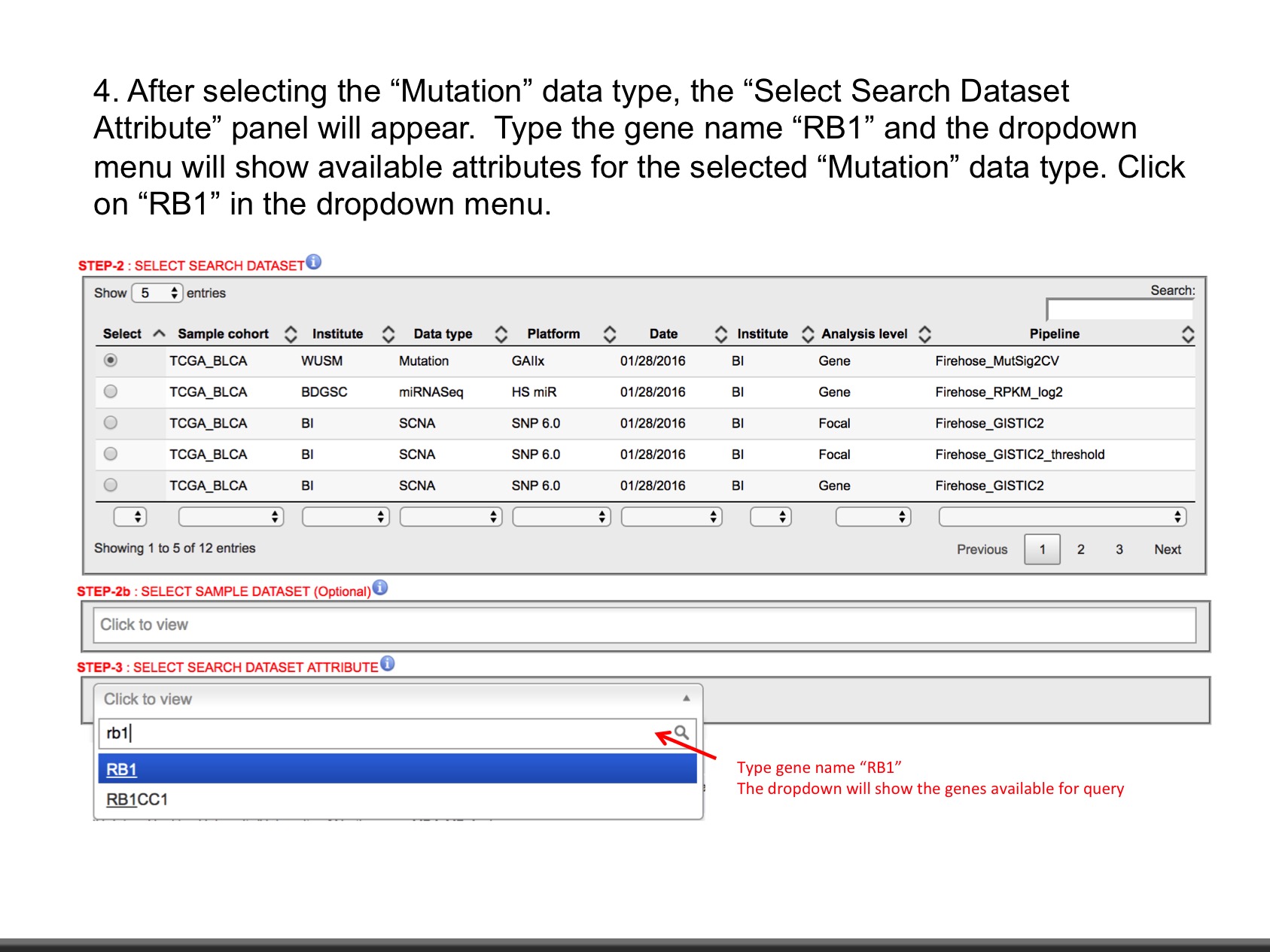
- Mutation (Level: Site, Gene) : Mutation calls for each participant
- Methylation (Level: Site, Gene) : Calculated beta values mapped to genome, per sample
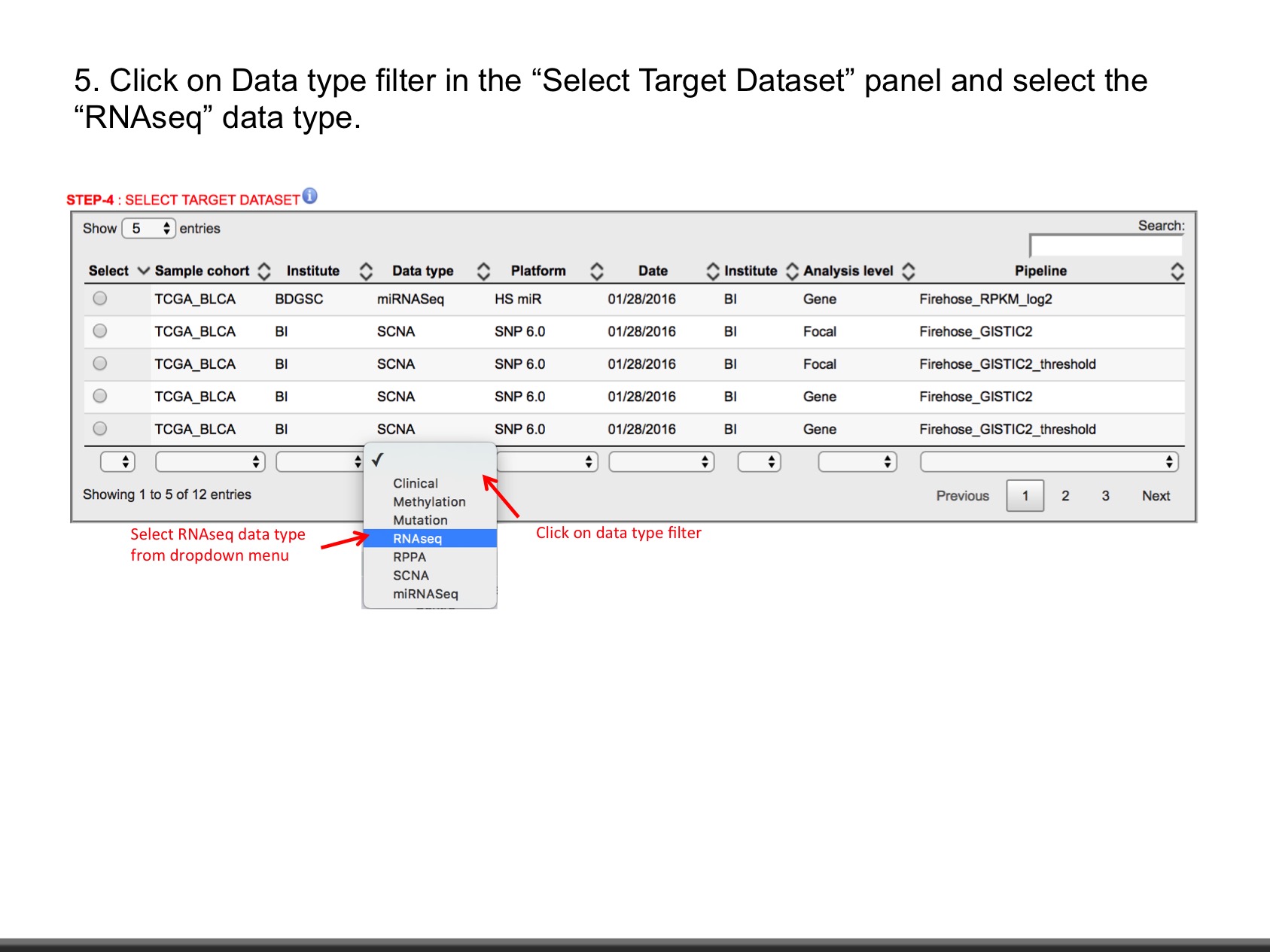
- RNAseq (Level: Gene, Isoform) : The normalized expression signal of individual Gene or isoforms (transcripts), per sample
- Expression Microarray (Level : Gene) : Normalized mRNA expression for each gene, per sample
- RPPA (Level: Analyte, Gene) : Normalized protein expression for each gene, per sample
- Proteomics (Level: Gene) : Average log-ratio of sample reporter-ion to common reference of peptide ions associated with the gene in acquisitions from a specific biological Sample (Unshared Log Ratio-Average log-ratio of sample reporter-ion to common reference of peptide ions of unshared peptides only associated with the gene in acquisitions from a specific biological sample). While, Spectral Counts represnts number of spectra matched to peptides associated with the gene in acquisitions from a specific biological sample (Unshared Spectral Counts -Number of spectra matched to unshared peptides associated with the gene in acquisitions from a specific biological sample).
- Phospho-Proteomics (Level: Site) : Average log-ratio of sample reporter-ion to common reference of peptide ions associated with phosphorylated site combinations in acquisitions from a specific biological sample (CDAP Protein Report).
- Glyco-Proteomics (Level: Site) : Average log-ratio of sample reporter-ion to common reference of peptide ions associated with deglycosylated N-glycosylation site combinations in acquisitions from a specific biological sample (CDAP Protein Report).
| Cancer Type | Cancer ID | Samples | Permission | Link |
|---|---|---|---|---|
| Adrenocortical carcinoma | ACC | 92 | Y | TCGA-Link |
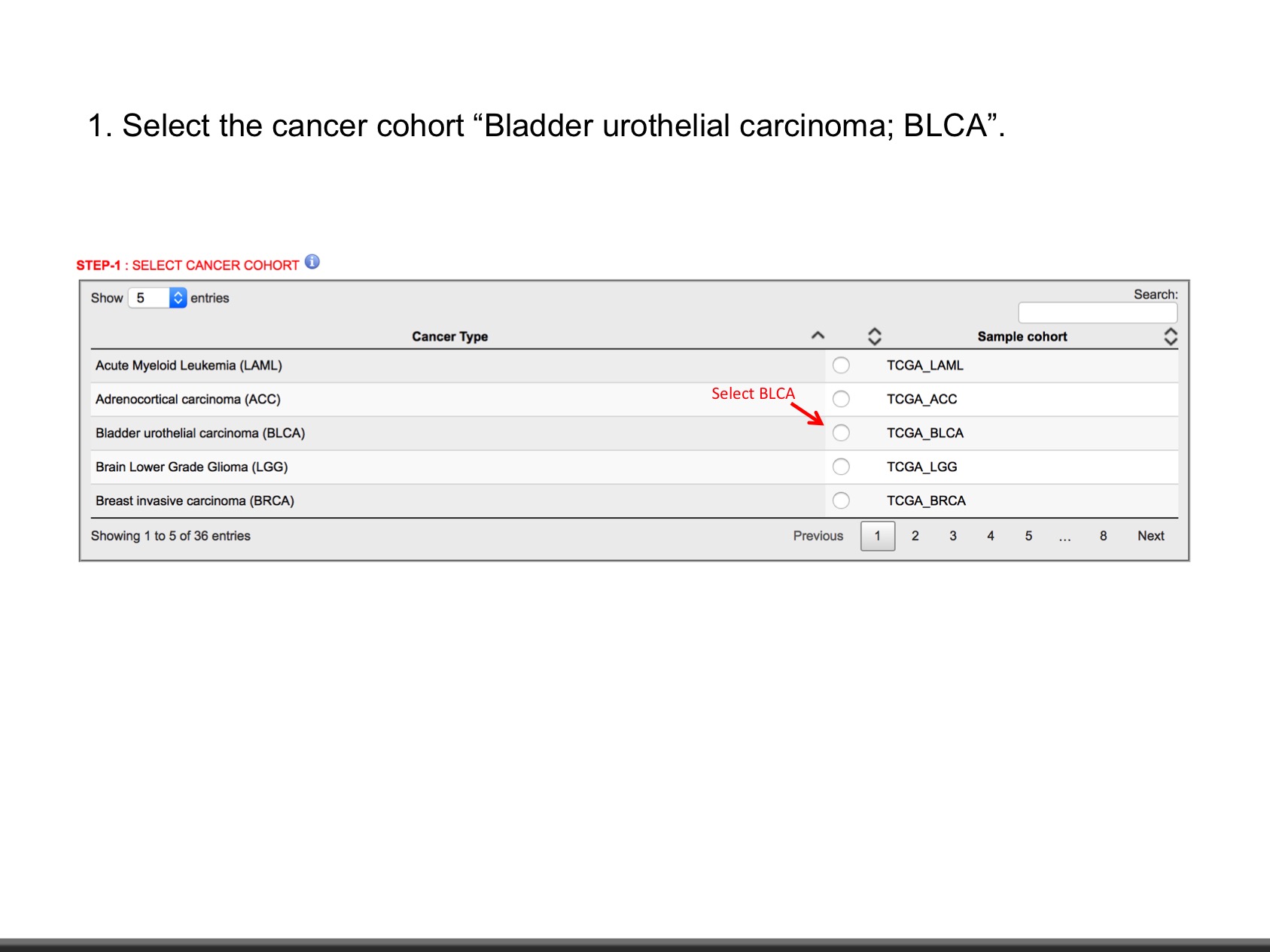
| Bladder urothelial carcinoma | BLCA | 412 | Y | TCGA-Link |
| Breast invasive carcinoma | BRCA | 1097 | Y | TCGA-Link CPTAC-Link |
| Cervical and endocervical cancers | CESC | 307 | Y | TCGA-Link |
| Cholangiocarcinoma | CHOL | 45 | Y | TCGA-Link |
| Colorectal adenocarcinoma | COADREAD | 629 | Y | TCGA-Link CPTAC-Link |
| Lymphoid Neoplasm Diffuse Large B-cell Lymphoma | DLBC | 48 | Y | TCGA-Link |
| Esophageal carcinoma | ESCA | 185 | Y | TCGA-Link |
| Glioblastoma multiforme | GBM | 595 | Y | TCGA-Link |
| Glioma | GBMLGG | 1110 | Y | TCGA-Link |
| Head and Neck squamous cell carcinoma | HNSC | 528 | Y | TCGA-Link |
| Kidney Chromophobe | KICH | 113 | Y | TCGA-Link |
| Pan-kidney cohort | KIPAN | 941 | Y | TCGA-Link |
| Kidney renal clear cell carcinoma | KIRC | 537 | Y | TCGA-Link |
| Kidney renal papillary cell carcinoma | KIRP | 291 | Y | TCGA-Link |
| Acute Myeloid Leukemia | LAML | 200 | Y | TCGA-Link |
| Brain Lower Grade Glioma | LGG | 515 | Y | TCGA-Link |
| Liver hepatocellular carcinoma | LIHC | 377 | Y | TCGA-Link |
| Lung adenocarcinoma | LUAD | 522 | Y | TCGA-Link |
| Lung squamous cell carcinoma | LUSC | 504 | Y | TCGA-Link |
| Mesothelioma | MESO | 87 | Y | TCGA-Link |
| Ovarian serous cystadenocarcinoma | OV | 591 | Y | TCGA-Link CPTAC-Link |
| Pancreatic adenocarcinoma | PAAD | 185 | Y | TCGA-Link |
| Pheochromocytoma and Paraganglioma | PCPG | 179 | Y | TCGA-Link |
| Prostate adenocarcinoma | PRAD | 499 | Y | TCGA-Link |
| Sarcoma | SARC | 261 | Y | TCGA-Link |
| Skin Cutaneous Melanoma | SKCM | 470 | Y | TCGA-Link |
| Stomach adenocarcinoma | STAD | 443 | Y | TCGA-Link |
| Stomach and Esophageal carcinoma | STES | 628 | Y | TCGA-Link |
| Testicular Germ Cell Tumors | TGCT | 134 | Y | TCGA-Link |
| Thyroid carcinoma | THCA | 503 | Y | TCGA-Link |
| Thymoma | THYM | 124 | Y | TCGA-Link |
| Uterine Corpus Endometrial Carcinoma | UCEC | 548 | Y | TCGA-Link |
| Uterine Carcinosarcoma | UCS | 57 | Y | TCGA-Link |
| Uveal Melanoma | UVM | 80 | Y | TCGA-Link |
| CellLine Colorectal adenocarcinoma | CL_COADREAD | 44 | N | - |
- Download Manual here Manual version 1.1
- Question? Please join the new User Forum.
- Explore data as shown in slides.
We are happy to integrate your data into LinkedOmics portal.
- Please contact admin to share your data (linkedomics.zhanglab@gmail.com).
The tool best run with browser Chrome Version 56.0.2924.87+ (download). If you are using Safari then enable the hidden Develop menu in Safari: Pull down the “Safari” menu and choose “Preferences” Click on the “Advanced” tab. Check the box next to “Show Develop menu in menu bar”. Click Develop menu and select Chrome/Firefox as browser.